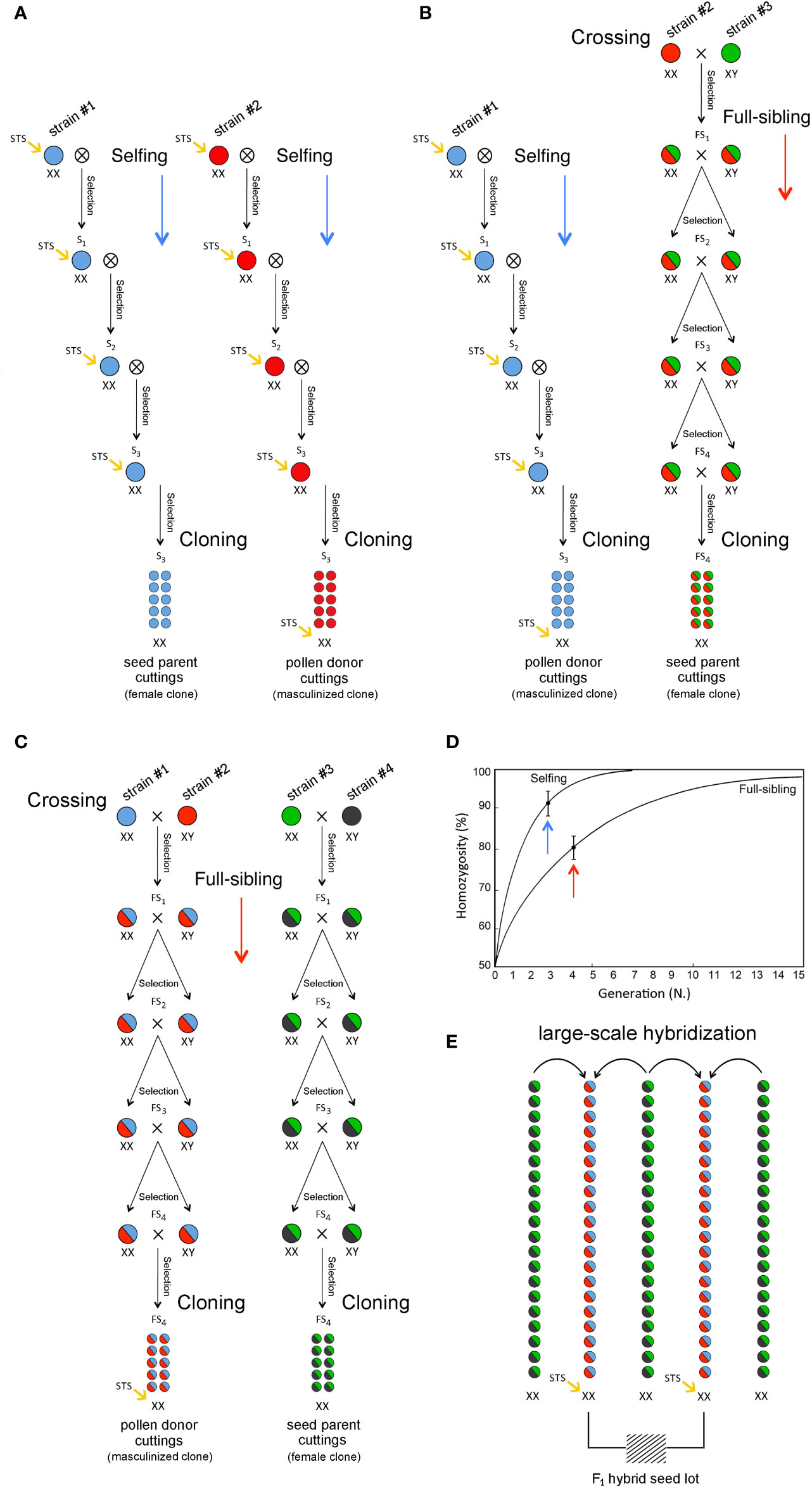
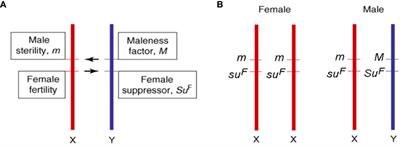
BobChronic6505
Active member
Ok so I'm not really sure how to title this post but I have a few general questions about breeding that I hope the pros here can help with.
So if i understand correctly, F2 generation follows mendels law of separation, where all the recessive traits show themselves....so are these recessive traits now considered dominate? If so, is it only for the F2 generation?
I made some F2s and now I'm sprouting them....I want to find a male and breed it back to one of the same F1s. Would this be considered an IBL or just a regular backcross? Also, let's say I do the F2 into F1. Are those dominate traits, that may only show during the F2, are they now going to be dominate traits in the resulting seeds? Are those seeds considered F3? Or is F3 only for putting the F2 to F2?
Also has anyone here bred some landrance shit into a fairly indica dom? Realistically, are those seeds going to be any good? Or are they going to be mutants? I know people do this all the time but I've always been skeptical about buying those kinds of seeds.
Thanks in advance
So if i understand correctly, F2 generation follows mendels law of separation, where all the recessive traits show themselves....so are these recessive traits now considered dominate? If so, is it only for the F2 generation?
I made some F2s and now I'm sprouting them....I want to find a male and breed it back to one of the same F1s. Would this be considered an IBL or just a regular backcross? Also, let's say I do the F2 into F1. Are those dominate traits, that may only show during the F2, are they now going to be dominate traits in the resulting seeds? Are those seeds considered F3? Or is F3 only for putting the F2 to F2?
Also has anyone here bred some landrance shit into a fairly indica dom? Realistically, are those seeds going to be any good? Or are they going to be mutants? I know people do this all the time but I've always been skeptical about buying those kinds of seeds.
Thanks in advance